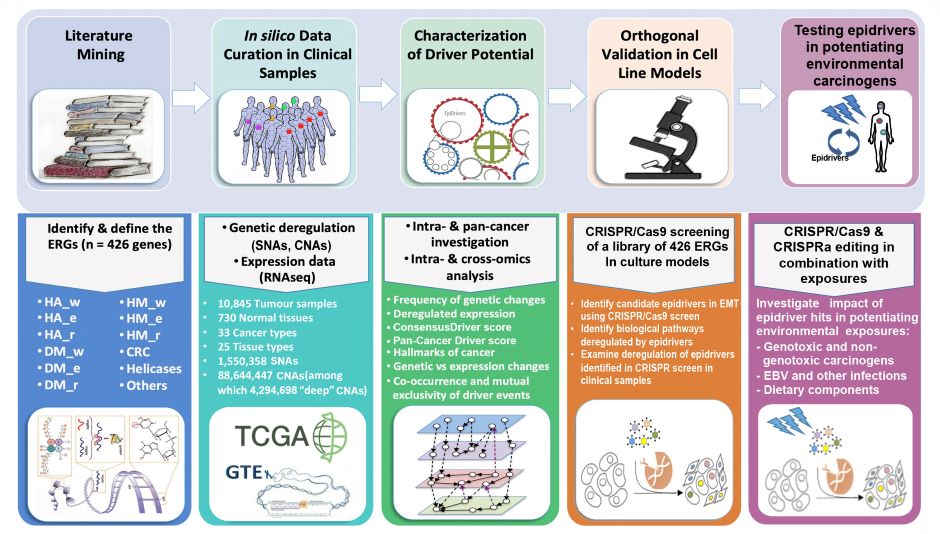
The main goal of EpiDRIVERS, a project of the Epigenomics and Mechanisms Branch (EGM), is to investigate whether epigenetic regulator genes (ERGs), when disrupted through genetic or non-mutational mechanisms, act as drivers (“epidrivers”) in cancer development and confer a cancer cell phenotype (through potential synergy with environmental exposure). To this end, EGM developed and tested a conceptual framework (involving a pan-cancer in silico genomic and experimental strategy) to identify and orthogonally validate functionally important epidriver genes through a novel systematic approach that uses the strengths of state-of-the-art genome-editing screens (Figure 1).
Figure 1. General pan-cancer genomic and experimental strategy for identifying and characterizing epigenetic driver genes and their environmental determinants. A five-stage approach is adopted to identify and assess epigenetic regulator genes (ERGs) with driver potential: (1) comprehensive literature mining, (2) in silico data curation in clinical samples, (3) modelling the driver potential of candidate genes, (4) using the CRISPR/Cas9 and CRISPR/dCas9 systems for orthogonal in vitro assessment of driver potential, and (5) characterizing the synergy between epidrivers and environmental exposures. Adapted from Halaburkova et al. (2020). © 2020 Halaburkova et al.; Published by Cold Spring Harbor Laboratory Press.
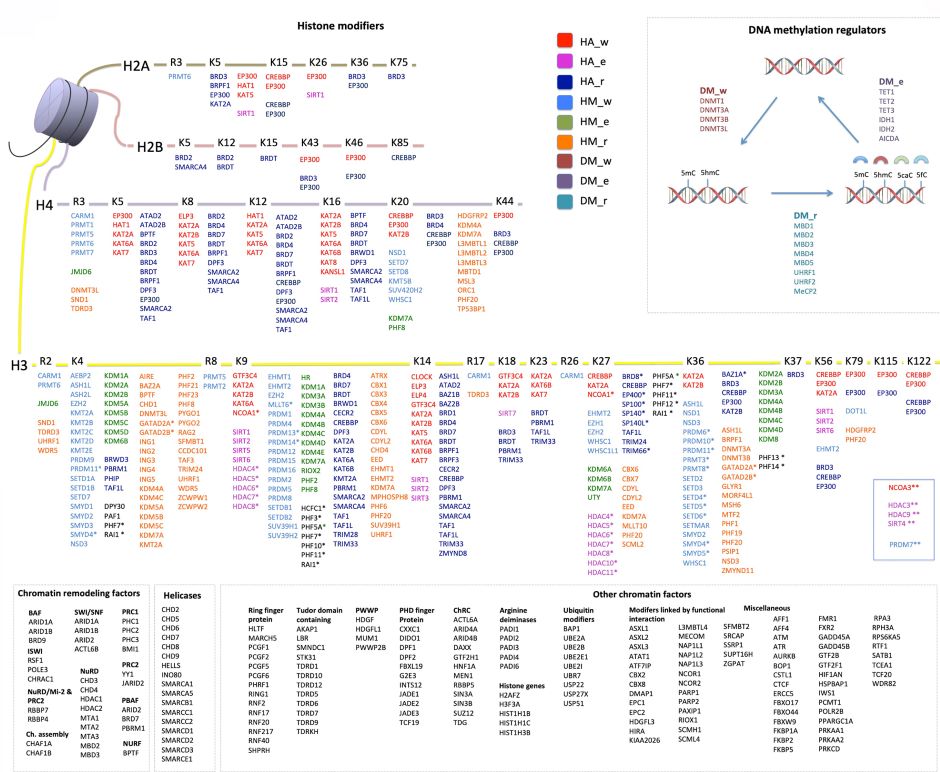
Epidemiological studies have uncovered robust associations between environmental and/or lifestyle factors and cancer risk; however, these studies are often limited by uncertain biological plausibility for the detected associations, which are often correlative in nature. Therefore, mechanistic studies may provide important contributions to understanding the causes of cancer, some of which (e.g. epigenetic deregulation) have emerged more recently. Consistent with this notion, one of the most remarkable findings of the international high-resolution cancer genome sequencing efforts is the high frequency of genetic alterations in ERGs (Figure 2) in common human cancer types.
Figure 2. A compendium of epigenetic regulator genes (ERGs) included in the study. This comprises 426 ERGs classified into the main categories: histone modifiers, chromatin remodellers, and DNA methylation regulators. Histone acetylation, histone methylation, and DNA methylation modifiers are each further stratified into “writers” (w), “editors” (e), and “readers” (r). The remaining ERGs were grouped into chromatin remodelling factors (ChRC), helicases, and other chromatin modifiers (some of which were further divided into subgroups based on function or their presence in molecular complexes). An asterisk (*) denotes the histone-modifying genes whose functions are not well characterized and which were therefore assigned based on ENCODE ChIP sequencing data; two asterisks (**) denote the histone-modifying genes without assignment of residues in the histone tails. From Halaburkova et al. (2020). © 2020 Halaburkova et al.; Published by Cold Spring Harbor Laboratory Press.
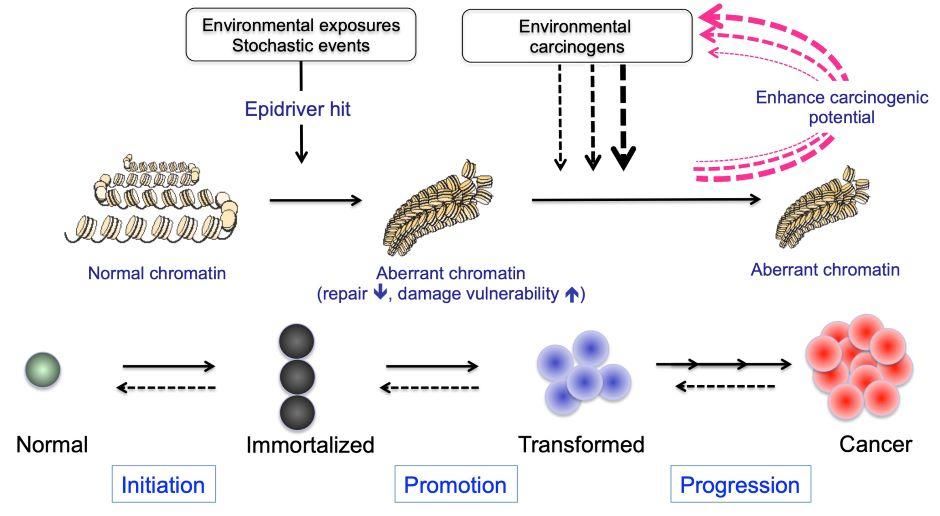
This high rate of genetic deregulation of ERGs constitutes a “genetic smoking gun” that epigenetic mechanisms lie at the very heart of cancer biology (Figure 3). However, much remains unknown about the impact of these changes on the expression and function of ERGs.
Figure 3. Hypothetical model of enhancing carcinogenic potential by chromatin changes induced by epidriver hits. Epidriver hits occurring in cells (as results of environmental exposure or stochastic events, such as those associated with ageing) might trigger or potentiate the carcinogenic effect of exposure to environmental carcinogens, leading to the development and progression of cancer.
This project builds on EGM’s previous work that provided important conceptual advances on the role of epigenetic regulators in chromatin-based processes, including transcription and DNA repair.
 View the image
View the image